HLA-A*02:01 binding "FLTGIGIITV" at 2.35Å resolution
Data provenance
Information sections
- Publication
- Peptide details
- Peptide neighbours
- Binding cleft pockets
- Chain sequences
- Downloadable data
- Data license
- Footnotes
Complex type
HLA-A*02:01
FLTGIGIITV
Species
Locus / Allele group
T-Cell Receptor optimized peptide skewing of the T-cell repertoire can enhance antigen targeting.
Altered peptide antigens that enhance T-cell immunogenicity have been used to improve peptide-based vaccination for a range of diseases. Although this strategy can prime T-cell responses of greater magnitude, the efficacy of constituent T-cell clonotypes within the primed population can be poor. To overcome this limitation, we isolated a CD8(+) T-cell clone (MEL5) with an enhanced ability to recognize the HLA A*0201-Melan A(27-35) (HLA A*0201-AAGIGILTV) antigen expressed on the surface of malignant melanoma cells. We used combinatorial peptide library screening to design an optimal peptide sequence that enhanced functional activation of the MEL5 clone, but not other CD8(+) T-cell clones that recognized HLA A*0201-AAGIGILTV poorly. Structural analysis revealed the potential for new contacts between the MEL5 T-cell receptor and the optimized peptide. Furthermore, the optimized peptide was able to prime CD8(+) T-cell populations in peripheral blood mononuclear cell isolates from multiple HLA A*0201(+) individuals that were capable of efficient HLA A*0201(+) melanoma cell destruction. This proof-of-concept study demonstrates that it is possible to design altered peptide antigens for the selection of superior T-cell clonotypes with enhanced antigen recognition properties.
Structure deposition and release
Data provenance
Publication data retrieved from PDBe REST API8 and PMCe REST API9
Other structures from this publication



Data provenance
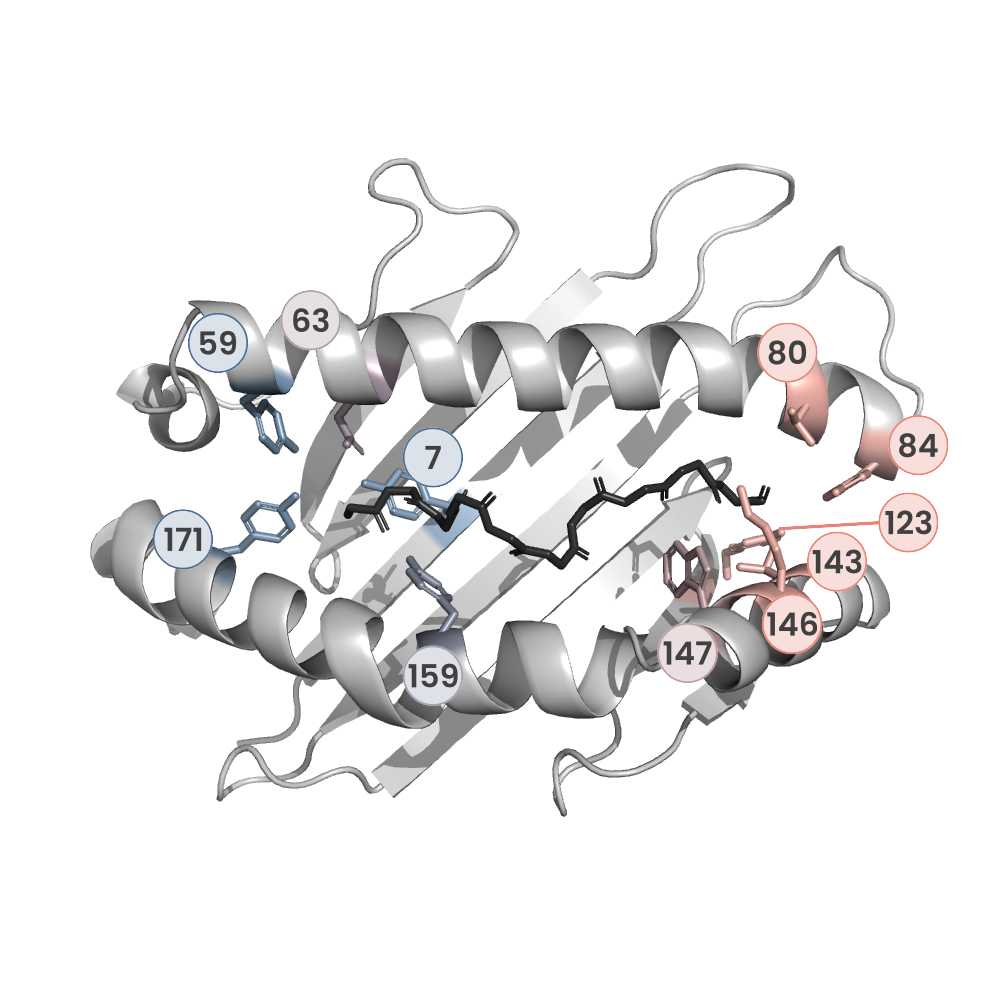
MHC:peptide complexes are visualised using PyMol. The peptide is superimposed on a consistent cutaway slice of the MHC binding cleft (displayed as a grey mesh) which best indicates the binding pockets for the P1/P5/PC positions (side view - pockets A, E, F) and for the P2/P3/PC-2 positions (top view - pockets B, C, D). In some cases peptides will use a different pocket for a specific peptide position (atypical anchoring). On some structures the peptide may appear to sterically clash with a pocket. This is an artefact of picking a standardised slice of the cleft and overlaying the peptide.
Peptide neighbours
|
P1
PHE
TYR59
GLU63
PHE33
TYR7
LYS66
TRP167
MET5
TYR171
TYR159
THR163
|
P10
VAL
LEU81
THR142
ASP77
TYR116
THR143
TYR84
THR80
LYS146
TRP147
TYR123
|
P2
LEU
LYS66
HIS70
VAL67
MET45
TYR99
TYR159
TYR7
GLU63
PHE9
|
P3
THR
LEU156
LYS66
HIS70
TYR99
TYR159
|
P4
GLY
TYR159
LYS66
|
P5
ILE
GLN155
LEU156
TYR159
ALA158
|
P6
GLY
ARG97
VAL152
GLN155
LEU156
HIS114
|
P7
ILE
ARG97
HIS70
TYR99
THR73
LEU156
|
P8
ILE
LYS146
THR73
VAL152
ASP77
ALA150
TRP147
ARG97
|
P9
THR
VAL76
LYS146
TRP147
THR73
ASP77
|
Colour key
Data provenance
Neighbours are calculated by finding residues with atoms within 5Å of each other using BioPython Neighboursearch module. The list of neighbours is then sorted and filtered to inlcude only neighbours where between the peptide and the MHC Class I alpha chain.
Colours selected to match the YRB scheme. [https://www.frontiersin.org/articles/10.3389/fmolb.2015.00056/full]

|
A Pocket
TYR159
THR163
TRP167
TYR171
MET5
TYR59
GLU63
LYS66
TYR7
|
B Pocket
ALA24
VAL34
MET45
GLU63
LYS66
VAL67
TYR7
HIS70
PHE9
TYR99
|
C Pocket
HIS70
THR73
HIS74
PHE9
ARG97
|
D Pocket
HIS114
GLN155
LEU156
TYR159
LEU160
TYR99
|
E Pocket
HIS114
TRP147
VAL152
LEU156
ARG97
|
F Pocket
TYR116
TYR123
THR143
LYS146
TRP147
ASP77
THR80
LEU81
TYR84
VAL95
|
Colour key
Data provenance
|
1. Beta 2 microglobulin
Beta 2 microglobulin
|
10 20 30 40 50 60
MIQRTPKIQVYSRHPAENGKSNFLNCYVSGFHPSDIEVDLLKNGERIEKVEHSDLSFSKD 70 80 90 WSFYLLYYTEFTPTEKDEYACRVNHVTLSQPKIVKWDRDM |
|
2. Class I alpha
HLA-A*02:01
IPD-IMGT/HLA
[ipd-imgt:HLA35266] |
10 20 30 40 50 60
GSHSMRYFFTSVSRPGRGEPRFIAVGYVDDTQFVRFDSDAASQRMEPRAPWIEQEGPEYW 70 80 90 100 110 120 DGETRKVKAHSQTHRVDLGTLRGYYNQSEAGSHTVQRMYGCDVGSDWRFLRGYHQYAYDG 130 140 150 160 170 180 KDYIALKEDLRSWTAADMAAQTTKHKWEAAHVAEQLRAYLEGTCVEWLRRYLENGKETLQ 190 200 210 220 230 240 RTDAPKTHMTHHAVSDHEATLRCWALSFYPAEITLTWQRDGEDQTQDTELVETRPAGDGT 250 260 270 FQKWAAVVVPSGQEQRYTCHVQHEGLPKPLTLRWEP |
|
3. Peptide
|
FLTGIGIITV
|
Data provenance
Sequences are retrieved via the Uniprot method of the RSCB REST API. Sequences are then compared to those derived from the PDB file and matched against sequences retrieved from the IPD-IMGT/HLA database for human sequences, or the IPD-MHC database for other species. Mouse sequences are matched against FASTA files from Uniprot. Sequences for the mature extracellular protein (signal petide and cytoplasmic tail removed) are compared to identical length sequences from the datasources mentioned before using either exact matching or Levenshtein distance based matching.
Downloadable data
Components
Data license
Footnotes
- Protein Data Bank Europe - Coordinate Server
- 1HHK - HLA-A*02:01 binding LLFGYPVYV at 2.5Å resolution - PDB entry for 1HHK
- Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. - PyMol CEALIGN Method - Publication
- PyMol - PyMol.org/pymol
- Levenshtein distance - Wikipedia entry
- Protein Data Bank Europe REST API - Molecules endpoint
- 3Dmol.js: molecular visualization with WebGL - 3DMol.js - Publication
- Protein Data Bank Europe REST API - Publication endpoint
- PubMed Central Europe REST API - Articles endpoint

This work is licensed under a Creative Commons Attribution 4.0 International License.