HLA-A*02:01 binding "SLFNTVATLY" at 2.00Å resolution
Data provenance
Information sections
- Publication
- Peptide details
- Peptide neighbours
- Binding cleft pockets
- Chain sequences
- Downloadable data
- Data license
- Footnotes
Complex type
HLA-A*02:01
SLFNTVATLY
Species
Locus / Allele group
Antigen processing influences HIV-specific cytotoxic T lymphocyte immunodominance.
Metal oxide heterostructures have gained huge attention in the energy storage applications due to their outstanding properties compared to pristine metal oxides. Herein, magnetic Fe2O3@SnO2 heterostructures were synthesized by the sol-gel electrospinning method at calcination temperatures of 450 and 600 °C. XRD line profile analysis indicated that fraction of tetragonal tin oxide phase compared to rhombohedral hematite was enhanced by increasing calcination temperature. FESEM images revealed that hexagonal nanoplatelets of Fe2O3 were hierarchically anchored on SnO2 hollow nanofibers. Optical band gap of heterogeneous structures was increased from 2.06 to 2.40 eV by calcination process. Vibrating sample magnetometer analysis demonstrated that increasing calcination temperature of the samples reduces saturation magnetization from 2.32 to 0.92 emu g-1. The Fe2O3@SnO2-450 and Fe2O3@SnO2-600 nanofibers as active materials coated onto Ni foams (NF) and their electrochemical performance were evaluated in three and two-electrode configurations in 3 M KOH electrolyte solution. Fe2O3@SnO2-600/NF electrode exhibits a high specific capacitance of 562.3 F g-1 at a current density of 1 A g-1 and good cycling stability with 92.8% capacitance retention at a high current density of 10 A g-1 after 3000 cycles in three-electrode system. The assembled Fe2O3@SnO2-600//activated carbon asymmetric supercapacitor device delivers a maximum energy density of 50.2 Wh kg-1 at a power density of 650 W kg-1. The results display that the Fe2O3@SnO2-600 can be a promising electrode material in supercapacitor applications.
Structure deposition and release
Data provenance
Publication data retrieved from PDBe REST API8 and PMCe REST API9
Other structures from this publication


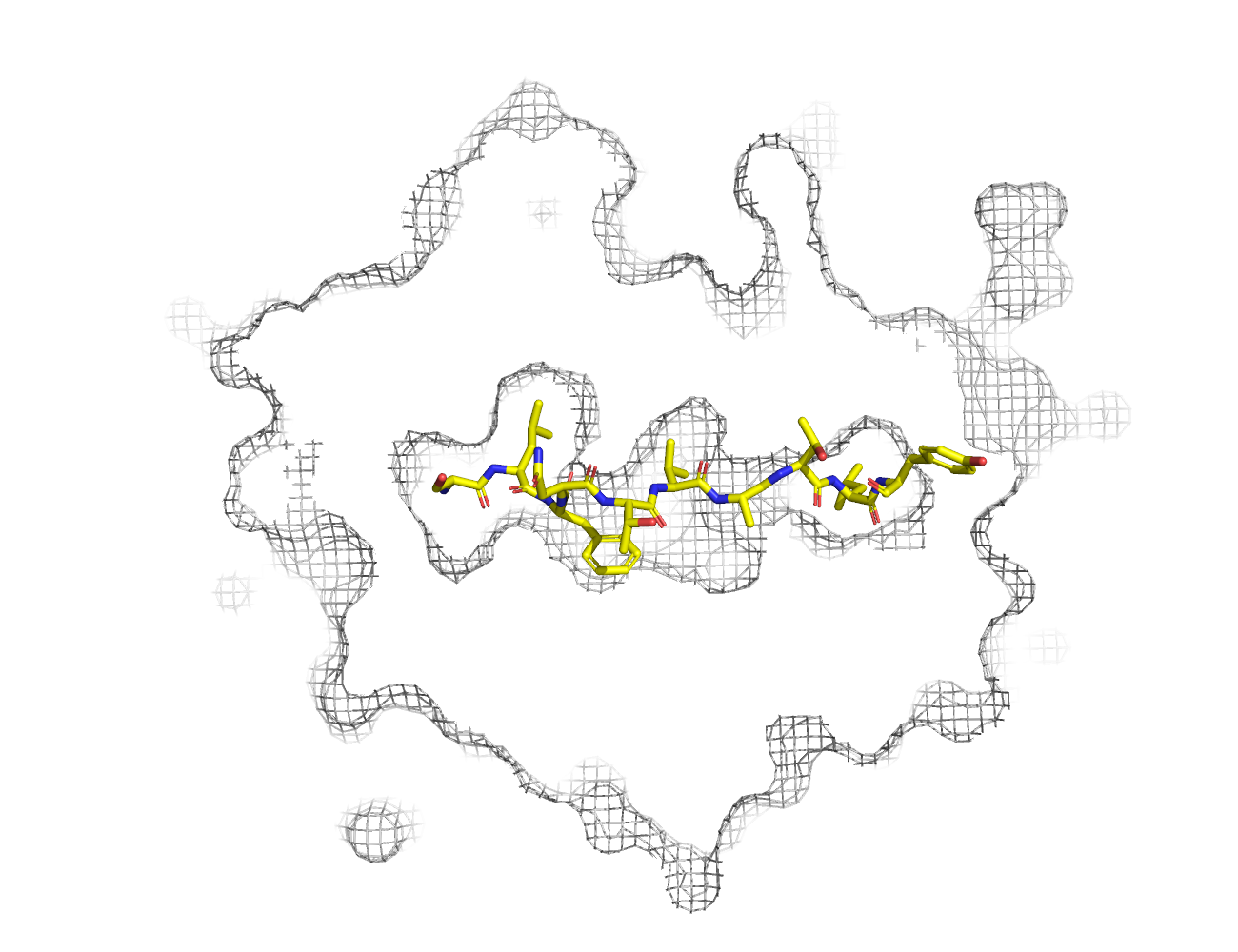
Data provenance
MHC:peptide complexes are visualised using PyMol. The peptide is superimposed on a consistent cutaway slice of the MHC binding cleft (displayed as a grey mesh) which best indicates the binding pockets for the P1/P5/PC positions (side view - pockets A, E, F) and for the P2/P3/PC-2 positions (top view - pockets B, C, D). In some cases peptides will use a different pocket for a specific peptide position (atypical anchoring). On some structures the peptide may appear to sterically clash with a pocket. This is an artefact of picking a standardised slice of the cleft and overlaying the peptide.
Peptide neighbours
|
P1
SER
TYR171
GLU63
LYS66
TYR159
TYR59
TRP167
PHE33
MET5
TYR7
|
P10
TYR
THR80
LYS146
ASP77
TYR84
|
P2
LEU
TYR159
PHE9
MET45
TYR7
TYR99
HIS70
GLU63
LYS66
VAL67
|
P3
PHE
TYR99
LYS66
TYR159
GLN155
LEU156
HIS70
|
P4
ASN
ARG65
LYS66
|
P5
THR
GLN155
|
P6
VAL
ALA69
THR73
HIS70
ARG97
|
P7
ALA
THR73
TRP147
ARG97
VAL152
ASP77
|
P8
THR
TRP147
VAL76
ASP77
THR73
|
P9
LEU
THR143
TRP147
ILE124
TYR116
TYR123
LYS146
ASP77
THR80
TYR84
LEU81
|
Colour key
Data provenance
Neighbours are calculated by finding residues with atoms within 5Å of each other using BioPython Neighboursearch module. The list of neighbours is then sorted and filtered to inlcude only neighbours where between the peptide and the MHC Class I alpha chain.
Colours selected to match the YRB scheme. [https://www.frontiersin.org/articles/10.3389/fmolb.2015.00056/full]


|
A Pocket
TYR159
THR163
TRP167
TYR171
MET5
TYR59
GLU63
LYS66
TYR7
|
B Pocket
ALA24
VAL34
MET45
GLU63
LYS66
VAL67
TYR7
HIS70
PHE9
TYR99
|
C Pocket
HIS70
THR73
HIS74
PHE9
ARG97
|
D Pocket
HIS114
GLN155
LEU156
TYR159
LEU160
TYR99
|
E Pocket
HIS114
TRP147
VAL152
LEU156
ARG97
|
F Pocket
TYR116
TYR123
THR143
LYS146
TRP147
ASP77
THR80
LEU81
TYR84
VAL95
|
Colour key
Data provenance
|
1. Beta 2 microglobulin
Beta 2 microglobulin
|
10 20 30 40 50 60
MIQRTPKIQVYSRHPAENGKSNFLNCYVSGFHPSDIEVDLLKNGERIEKVEHSDLSFSKD 70 80 90 WSFYLLYYTEFTPTEKDEYACRVNHVTLSQPKIVKWDRDM |
|
2. Class I alpha
HLA-A*02:01
IPD-IMGT/HLA
[ipd-imgt:HLA35266] |
10 20 30 40 50 60
GSHSMRYFFTSVSRPGRGEPRFIAVGYVDDTQFVRFDSDAASQRMEPRAPWIEQEGPEYW 70 80 90 100 110 120 DGETRKVKAHSQTHRVDLGTLRGYYNQSEAGSHTVQRMYGCDVGSDWRFLRGYHQYAYDG 130 140 150 160 170 180 KDYIALKEDLRSWTAADMAAQTTKHKWEAAHVAEQLRAYLEGTCVEWLRRYLENGKETLQ 190 200 210 220 230 240 RTDAPKTHMTHHAVSDHEATLRCWALSFYPAEITLTWQRDGEDQTQDTELVETRPAGDGT 250 260 270 FQKWAAVVVPSGQEQRYTCHVQHEGLPKPLTLRWE |
|
3. Peptide
|
SLFNTVATLY
|
Data provenance
Sequences are retrieved via the Uniprot method of the RSCB REST API. Sequences are then compared to those derived from the PDB file and matched against sequences retrieved from the IPD-IMGT/HLA database for human sequences, or the IPD-MHC database for other species. Mouse sequences are matched against FASTA files from Uniprot. Sequences for the mature extracellular protein (signal petide and cytoplasmic tail removed) are compared to identical length sequences from the datasources mentioned before using either exact matching or Levenshtein distance based matching.
Downloadable data
Components
Data license
Footnotes
- Protein Data Bank Europe - Coordinate Server
- 1HHK - HLA-A*02:01 binding LLFGYPVYV at 2.5Å resolution - PDB entry for 1HHK
- Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. - PyMol CEALIGN Method - Publication
- PyMol - PyMol.org/pymol
- Levenshtein distance - Wikipedia entry
- Protein Data Bank Europe REST API - Molecules endpoint
- 3Dmol.js: molecular visualization with WebGL - 3DMol.js - Publication
- Protein Data Bank Europe REST API - Publication endpoint
- PubMed Central Europe REST API - Articles endpoint

This work is licensed under a Creative Commons Attribution 4.0 International License.